dMDT Interaction Network
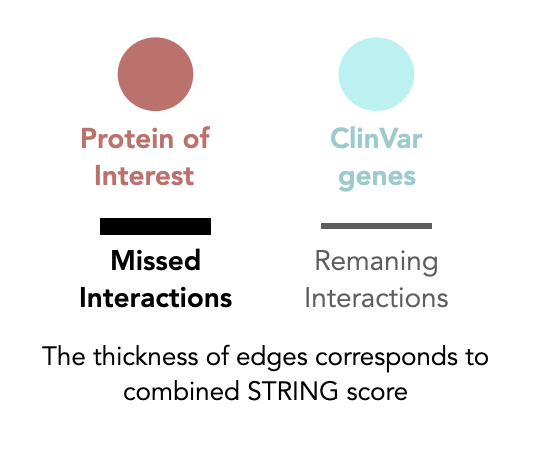
Protein interaction losses due to MDT switch
| Gene Name | Interaction Lost | Lost Domain (Region) | Lost Domain - Domain Interaction |
|---|---|---|---|
| MAP2K4 | MAP2K4-NFKB1 | Pkinase(104-367) | Pkinase(104-367) - Ank |
| MAP2K4 | MAP2K4-MAPK13 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAPK12 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAPK14 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-RIPK1 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP3K8 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-AKT1 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-PAK1 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP2K1 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-PAK2 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-ROCK2 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-PRKCD | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAPK11 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP3K2 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MINK1 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP3K5 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAPK10 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAPK8 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP3K3 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP3K7 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP3K4 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP2K7 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP3K1 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-ROCK1 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAPK9 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-MAP3K12 | Pkinase(104-367) | Pkinase(104-367) - Pkinase |
| MAP2K4 | MAP2K4-RAC3 | Pkinase(104-367) | Pkinase(104-367) - Ras |
| MAP2K4 | MAP2K4-RAC1 | Pkinase(104-367) | Pkinase(104-367) - Ras |
| MAP2K4 | MAP2K4-CASP10 | Pkinase(104-367) | Pkinase(104-367) - DED |
| MAP2K4 | MAP2K4-FADD | Pkinase(104-367) | Pkinase(104-367) - DED |
| MAP2K4 | MAP2K4-FLT4 | Pkinase(104-367) | Pkinase(104-367) - I-set |
| MAP2K4 | MAP2K4-MAP3K9 | Pkinase(104-367) | Pkinase(104-367) - Pkinase_Tyr |
Network Density Score: %
The local network of this protein has more interaction than % of all proteins in the proteome.
Statistics on dMDT
*This transcript has been found as a dMDT in these diseases
Samples showing lost interactions due to MDT switch